Methicillin Resistant Staphylococcus aureus (MRSA) Isolated from Skin Infections in Poultry (Bumble Foot)
Abstract
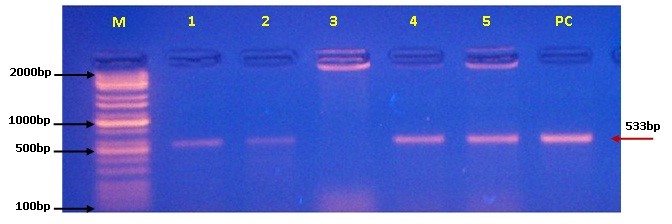
This study was aimed to isolate and identify of MRSA from infected skin of poultry, and testing the susceptibility of the isolates to different antibiotics especially methicillin, and confirmation of mecA gene encoded to methicillin resistance. Therefore, a total of 50 skin swabs were collected from the suspected poultry foot cases at different farm in Wasit province / Iraq, during November (2021) to January (2022). The swab samples were grown and purified, and then, the isolates were tested biochemically and phenotypically and subjected finally to antimicrobial sensitivity testing and molecular confirmation using conventional PCR assay. Examination of suspected S. aureus isolates revealed that the number of grown and fermented isolates on Mannitol salt agar (MSA) was 48%, and 12% for non-fermented MSA isolates; while, 40% of isolates were not grown on MSA. The colonies of suspected S. aureus were appeared on MSA as large, convex, shiny to opaque, circular, and golden-yellowish color; while on blood agar, colonies had signif, rounded, smooth, and grayish appearance. All grown and fermented isolates were showed positive reactivity to Catalase (100%) and Bound Coagulase (87.5%) test
References
Manandhar, S., Singh, A., and Shrivastava, N. (2021). Phenotypic and genotypic characterization of biofilm producing clinical coagulase negative staphylococci from Nepal and their antibiotic susceptibility pattern. Ann Clinic Microbiol Antimicrobials, 20(1), 1-11.
Peton, V., and Le Loir, Y. (2014). Staphylococcus aureus in veterinary medicine. Infection, Genetics and Evolution, 21, 602-615.
Mamza, S. A., Geidam, Y. A., Mshelia, G. D., Egwu, G. O., and Gulani, I. (2016). Morphological and biochemical characterization of Staphylococci isolated from food-producing animals in Northern Nigeria. Direct Res. J, 1(1), 1-8.
Pietrocola, G., Nobile, G., Rindi, S., and Speziale, P. (2017). Staphylococcus aureus manipulates innate immunity through own and host-expressed proteases. Frontiers in cellular and infection microbiology, 7, 166.
Singh, V., and Phukan, U. J. (2019). Interaction of host and Staphylococcus aureus protease-system regulates virulence and pathogenicity. Medical Microbiology and Immunology, 208(5), 585-607.
Gordon, C. P., Williams, P., and Chan, W. C. (2013). Attenuating Staphylococcus aureus virulence gene regulation: a medicinal chemistry perspective. Journal of medicinal chemistry, 56(4), 1389-1404.
Foster, T. J. (2017). Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS microbiology reviews, 41(3), 430-449.
Gnanamani, A., Hariharan, P., and Paul-Satyaseela, M. (2017). Staphylococcus aureus: Overview of bacteriology, clinical diseases, epidemiology, antibiotic resistance and therapeutic approach. Frontiers in Staphylococcus aureus, 4(28), 10-5772.
Lee, A. S., De Lencastre, H., Garau, J., Kluytmans, J., Malhotra-Kumar, S., Peschel, A., and Harbarth, S. (2018). Methicillin-resistant Staphylococcus aureus. Nature reviews Disease primers, 4(1), 1-23.
El-Sayed, A., Aleya, L., and Kamel, M. (2021). The link among microbiota, epigenetics, and disease development. Environmental Science and Pollution Research, 28(23), 28926-28964.
Blair, J. (2013). Bumblefoot: a comparison of clinical presentation and treatment of pododermatitis in rabbits, rodents, and birds. Veterinary Clinics: Exotic Animal Practice, 16(3), 715-735.
Jacob, F. G., Baracho, M. D. S., and Salgado, D. D. A. (2016). The use of infrared thermography in the identification of pododermatitis in broilers. Engenharia Agrícola, 36, 253-259.
Miesle, J. (2021). Pododermatitis (Bumblefoot): Diagnosis, Treatment, and Resolution. Academia, 2021, 1-42.
Saksangawong, C., Padungtod, P., Pichpol, D., and Chanayath, N. (2005). Comparison of anticoagulants used in rabbit plasma preparation for Staphylococcus aureus coagulase test. Sattawaphaetthayasan.
Monica, C. (2006). District laboratory practice in tropical countries. Cambridge University Press, http://localhost:8080/xmlui/handle/123456789/3227.
Idrees, M., Sawant, S., Karodia, N., and Rahman, A. (2021). Staphylococcus aureus biofilm: Morphology, genetics, pathogenesis and treatment strategies. International Journal of Environmental Research and Public Health, 18(14), 7602.
Konuku, S., Rajan, M. M., and Muruhan, S. (2012). Morphological and biochemical characteristics and antibiotic resistance pattern of Staphylococcus aureus isolated from grapes. International Journal of Nutrition, Pharmacology, Neurological Diseases, 2(1), 70.
Wang, L. J., Yang, X., Qian, S. Y., Dong, F., and Song, W. Q. (2020). Identification of hemolytic activity and hemolytic genes of Methicillin-resistant Staphylococcus aureus isolated from Chinese children. Chinese medical journal, 133 (01), 88-90
Doulgeraki, A. I., Di Ciccio, P., Ianieri, A., and Nychas, G. J. E. (2017). Methicillin-resistant food-related Staphylococcus aureus: A review of current knowledge and biofilm formation for future studies and applications. Research in microbiology, 168(1), 1-15.
Kirby, W. M., Bauer, A. M., Sherris, J. C., and Turck, M. (1966). Antibiotic sensitivity testing by a standardized disc diffusion method. Am J Clin Pathol, 45(4), 493-496.
Butaye, P., Argudín, M. A., and Smith, T. C. (2016). Livestock-associated MRSA and its current evolution. Current Clinical Microbiology Reports, 3(1), 19-31.
Kavanagh, K. T., Abusalem, S., and Calderon, L. E. (2017). The incidence of MRSA infections in the United States: is a more comprehensive tracking system needed?. Antimicrobial Resistance and Infection Control, 6(1), 1-6.
de Vos, A. S., De Vlas, S. J., Lindsay, J. A., Kretzschmar, M. E., and Knight, G. M. (2021). Understanding MRSA clonal competition within a UK hospital; the possible importance of density dependence. Epidemics, 37, 100511.
Vestergaard, M., Frees, D., and Ingmer, H. (2019). Antibiotic resistance and the MRSA problem. Microbiology spectrum, 7(2), 7-2.
Javed, M. U., Ijaz, M., Fatima, Z., Anjum, A. A., Aqib, A. I., Ali, M. M., and Ghaffar, A. (2021). Frequency and Antimicrobial Susceptibility of Methicillin and Vancomycin-Resistant Staphylococcus aureus from Bovine Milk. Pakistan Veterinary Journal, 41(4).
Harkins, C. P., Pichon, B., Doumith, M., Parkhill, J., Westh, H., Tomasz, A., and Holden, M. T. (2017). Methicillin-resistant Staphylococcus aureus emerged long before the introduction of methicillin into clinical practice. Genome biology, 18(1), 1-11.
Samsudin, N. I. P., Lee, H. Y., Chern, P. E., Ng, C. T., Panneerselvam, L., Phang, S. Y., and Mahyudin, N. A. (2018). In vitro antibacterial activity of crude medicinal plant extracts against ampicillin+ penicillin-resistant Staphylococcus aureus. International Food Research Journal, 25(2).
Craft, K. M., Nguyen, J. M., Berg, L. J., and Townsend, S. D. (2019). Methicillin-resistant Staphylococcus aureus (MRSA): antibiotic-resistance and the biofilm phenotype. MedChemComm, 10(8), 1231-1241.
Lima, L. M., da Silva, B. N. M., Barbosa, G., and Barreiro, E. J. (2020). β-lactam antibiotics: An overview from a medicinal chemistry perspective. European Journal of Medicinal Chemistry, 208, 112829.
Bilyk, B. L., Panchal, V. V., Tinajero-Trejo, M., Hobbs, J. K., and Foster, S. J. (2022). An Interplay of Multiple Positive and Negative Factors Governs Methicillin Resistance in Staphylococcus aureus. Microbiology and Molecular Biology Reviews, e00159-21.
Bai, B. P. (2019). Study of Methicillin Beta Lactam Antibiotic with Penicillin Binding Protein 2A. Life Science Informatics Publications, 5 (2), 642-658.
Urushibara, N., Aung, M. S., Kawaguchiya, M., and Kobayashi, N. (2020). Novel staphylococcal cassette chromosome mec (SCC mec) type XIV (5A) and a truncated SCC mec element in SCC composite islands carrying speG in ST5 MRSA in Japan. Journal of Antimicrobial Chemotherapy, 75(1), 46-50.
Bloom, D. E., Black, S., Salisbury, D., and Rappuoli, R. (2018). Antimicrobial resistance and the role of vaccines. Proceedings of the National Academy of Sciences, 115(51), 12868-12871.
Abushaheen, M. A., Fatani, A. J., Alosaimi, M., Mansy, W., George, M., Acharya, S., and Jhugroo, P. (2020). Antimicrobial resistance, mechanisms and its clinical significance. Disease-a-Month, 66(6), 100971.
Freitas Ribeiro, L., Akira Sato, R., de Souza Pollo, A., Marques Rossi, G. A., and do Amaral, L. A. (2020). Occurrence of methicillin-resistant Staphylococcus spp. on Brazilian dairy farms that produce unpasteurized cheese. Toxins, 12(12), 779.
Sader, H. S., Rhomberg, P. R., Doyle, T. B., Flamm, R. K., and Mendes, R. E. (2018). Evaluation of the revised ceftaroline disk diffusion breakpoints when testing a challenge collection of methicillin-resistant Staphylococcus aureus isolates. Journal of clinical microbiology, 56(12), e00777-18.
Wilcox, M., Al-Obeid, S., Gales, A., Kozlov, R., Martínez-Orozco, J. A., Rossi, F., and Blondeau, J. (2019). Reporting elevated vancomycin minimum inhibitory concentration in methicillin-resistant Staphylococcus aureus: consensus by an International Working Group. Future microbiology, 14(4), 345-352.
Ghumman, N. Z., Ijaz, M., Ahmed, A., Javed, M. U., Muzammil, I., and Raza, A. (2022). Evaluation of Methicillin Resistance in Field Isolates of Staphylococcus aureus: An Emerging Issue of Indigenous Bovine Breeds. Pakistan J. Zool., 2022, 1-12.
Al-Khafaji, A. N. (2018). Isolation and identification of methicillin resistance Staphylococcus aureus and detection their ability to the production of virulence factors. Journal of University of Babylon for Pure and Applied Sciences, 26(8), 100-111.
Masimen, M. A. A., Harun, N. A., Maulidiani, M., and Ismail, W. I. W. (2022). Overcoming Methicillin-Resistance Staphylococcus aureus (MRSA) Using Antimicrobial Peptides-Silver Nanoparticles. Antibiotics, 11(7), 951.
Hidayat, Y. W., Widodo, A. D. W., and Dachan, Y. P. (2019). The antimicrobial effect of+ Oxivarea against methicillin resistance staphylococcus aureus and pseudomonas aeruginosa. Internet Journal of Microbiology, 16(1).
Nair, D., Shashindran, N., Kumar, A., Vinodh, V., Biswas, L., and Biswas, R. (2021). Comparison of phenotypic MRSA detection methods with PCR for mecA gene in the background of emergence of oxacillin-susceptible MRSA. Microbial Drug Resistance, 27(9), 1190-1194.
Noumi, E., Merghni, A., Alreshidi, M., Del Campo, R., Adnan, M., Haddad, O., and Snoussi, M. (2020). Phenotypic and genotypic characterization with MALDI-TOF-MS based identification of Staphylococcus spp. isolated from Mobile phones with their antibiotic susceptibility, biofilm formation, and adhesion properties. International Journal of Environmental Research and Public Health, 17(11), 3761.
Adekunle, O. C., Bolaji, O. S., Olalekan, A. O., and Oyakeye, T. O. (2021). Identification of Methicillin Resistance Staphylococcus aureus From Clinical Samples and Environments of a General Hospital in Osogbo. Journal of Applied Sciences and Environmental Management, 25(8), 1409-1414.
Lord, J., Millis, N., Jones, R. D., Johnson, B., and Kania, S. A. (2022). Patterns of antimicrobial, multidrug and methicillin resistance among Staphylococcus spp. isolated from canine specimens submitted to a diagnostic laboratory in Tennessee, USA: A descriptive study. BMC Veterinary Research, 18(1), 1-16.
Iwamoto, K., Moriwaki, M., and Hide, M. (2019). Staphylococcus aureus in atopic dermatitis: Strain-specific cell wall proteins and skin immunity. Allergology Inter, 68(3), 309-315.
Slavetinsky, C. J., Hauser, J. N., Gekeler, C., Slavetinsky, J., Geyer, A., Kraus, A., and Peschel, A. (2022). Sensitizing Staphylococcus aureus to antibacterial agents by decoding and blocking the lipid flippase MprF. Elife, 11, e66376.
Bakr, M. E., Kashef, M. T., Hosny, A. E. D., and Ramadan, M. A. (2022). Effect of spdC gene expression on virulence and antibiotic resistance in clinical Staphylococcus aureus isolates. International Microbiology, 1-11.
Gorr, S. U., Brigman, H. V., Anderson, J. C., and Hirsch, E. B. (2020). The antimicrobial peptide DGL13K is active against resistant gram-negative bacteria and subinhibitory concentrations stimulate bacterial growth without causing resistance. bioRxiv.
Missiakas, D., and Winstel, V. (2021). Selective host cell death by Staphylococcus aureus: A strategy for bacterial persistence. Frontiers in Immunology, 11, 621733.